Division of Structural Biomolecular Science
Many complex biological phenomena are caused by the functions of small molecules and changes in the interactions between biomolecules, which are not directly regulated using genetic information. In the postgenomic era, to gain insight in such phenomena, we focus on the structures and interactions of natural products, glycans, and biomembranes. Unveiling the localization and dynamics of metabolites is also important because they are related to their functions. Recent advanced spectroscopic instruments and organic molecular probes are essential tools for the study of molecular mechanisms in biological phenomena. In the context of structural biomolecular sciences, which are based on organic chemistry, biochemistry, analytical chemistry, and structural biology, we study molecular mechanisms of life phenomena by combining the visualization of the metabolite dynamics, the identification of bioactive molecules, and the chemical synthesis of functional molecules.
Group Members
Division of Structural Biomolecular Science
General Manager/ Executive Researcher
Tohru YAMAGAKI
Specially Appointed Manager
Keiko SHIMAMOTO
Senior Researcher
Kaoru NOMURA
Appointed Researcher
Manabu HORIKAWA
Researcher
Erisa HARADA
Researcher
Kohki FUJIKAWA
Researcher
Shoko MORI
Technical Researcher
Tsukiho OSAWA
Research Projects
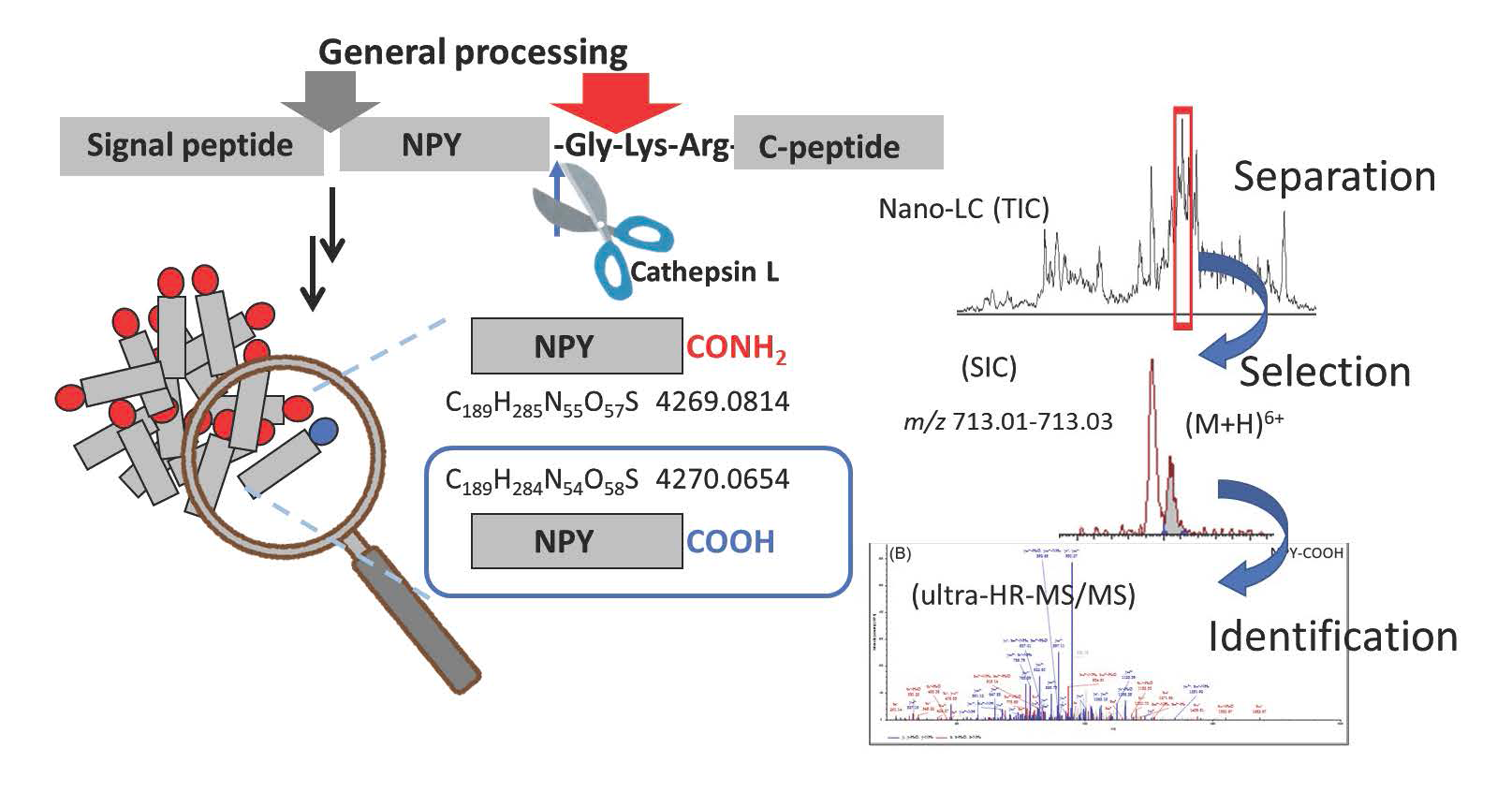
1. Detection of Minor Metabolites in the Neuropeptide Maturation Process
Neuropeptides are found in the central and peripheral nervous system, where they act as signaling molecules involved in endocrine functions, regulation of reproduction and feeding, learning and memory, and pain perception. Neuropeptides are expressed in vivo as large precursor peptides (prepropeptides). Subsequently, they undergo multistep posttranslational modifications in the cell, such as enzymatic digestion and amidation, to become mature peptides with physiological activity. Although the general maturation process of neuropeptides has been extensively studied, many aspects still remain unveiled because of the extremely small amounts of intermediary metabolites. MS is a very effective tool for tracing the maturation process of endogenous neuropeptides; however, the direct analysis of the processing products of large peptides having sizes of several kDa is difficult. Particularly challenging is the detection of a trace amount of non-amidated peptides in the presence of amidated peptides because their molecular weights differ by only 1 Da. During our studies on the maturation process of neuropeptide Y (NPY), by combining a pretreatment such as two-step gel filtration chromatography with nano-liquid chromatography (LC)/ultrahigh-resolution Orbitrap MS, we found that a small amount of non-amidated NPY-COOH is present in the mouse brain, which suggests that cathepsin L, a digestive enzyme that generates NPY-COOH, also functions here. We plan to examine metabolic peptides in detail to reveal the regulation of the neuropeptide production process and the physiological roles of minor metabolic peptides.
2. Elucidation of the Action Mechanisms of Lipid-related Molecules in the Presence of Biomembranes
All organisms have biomembranes, in which membrane proteins are responsible for the exchange of compounds and communication between cells. Although biomembranes are basically composed of a phospholipid bilayer, their compositions vary with the type of organelles or species. In addition to simple phospholipids, biomembranes also contain various conjugated lipids, such as glycolipids, as minor components.
The biological significance and roles of minor components remain poorly understood because the limitations of molecular biological approaches have hampered the elucidation of their functions. Thus, we have investigated the roles of lipidated biomolecules in biomembranes serving as specific reaction fields in biological events using both bioorganic chemical and physicochemical approaches.
Mechanisms of Chaperone- and Enzyme-like Activities of a Glycolipid
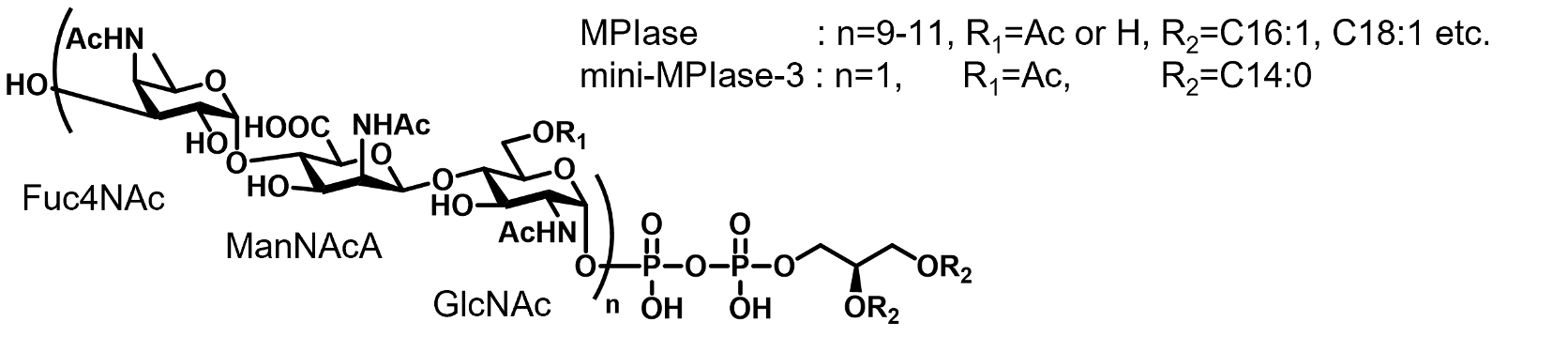
We previously identified MPIase (membrane protein integrase) as a novel glycolipid that is essential for membrane protein integration into the inner membrane of Escherichia coli. MPIase contains a glycan moiety consisting of approximately 10 repeating trisaccharide units and a pyrophospholipid. Because MPIase is the first known glycolipid that acts like an enzyme, we proposed the new term “glycolipozyme” to refer to this type of glycolipid. We studied the mechanism of the chaperone- and enzyme-like activities of MPIase. To date, our results can be summarized as follows:
(1) We successfully synthesized the minimal structure (mini-MPIase-3) required for MPIase to exhibit membrane integration activity. Additionally, we synthesized various MPIase analogs and identified the functional groups involved in the membrane integration activity by conducting structure-activity relationship studies.
(2) Using the synthesized analogs and natural MPIase, we analyzed the interactions between MPIase, a substrate protein, and membrane lipids by performing nuclear magnetic resonance (NMR), surface plasmon resonance (SPR), and fluorescence measurements.
(3) We identified the initial step in the MPIase biosynthetic pathway using the synthesized biosynthetic intermediates and substrate candidates of MPIase.
These studies allowed revealing the action mechanism of MPIase in the membrane integration process of substrate proteins.
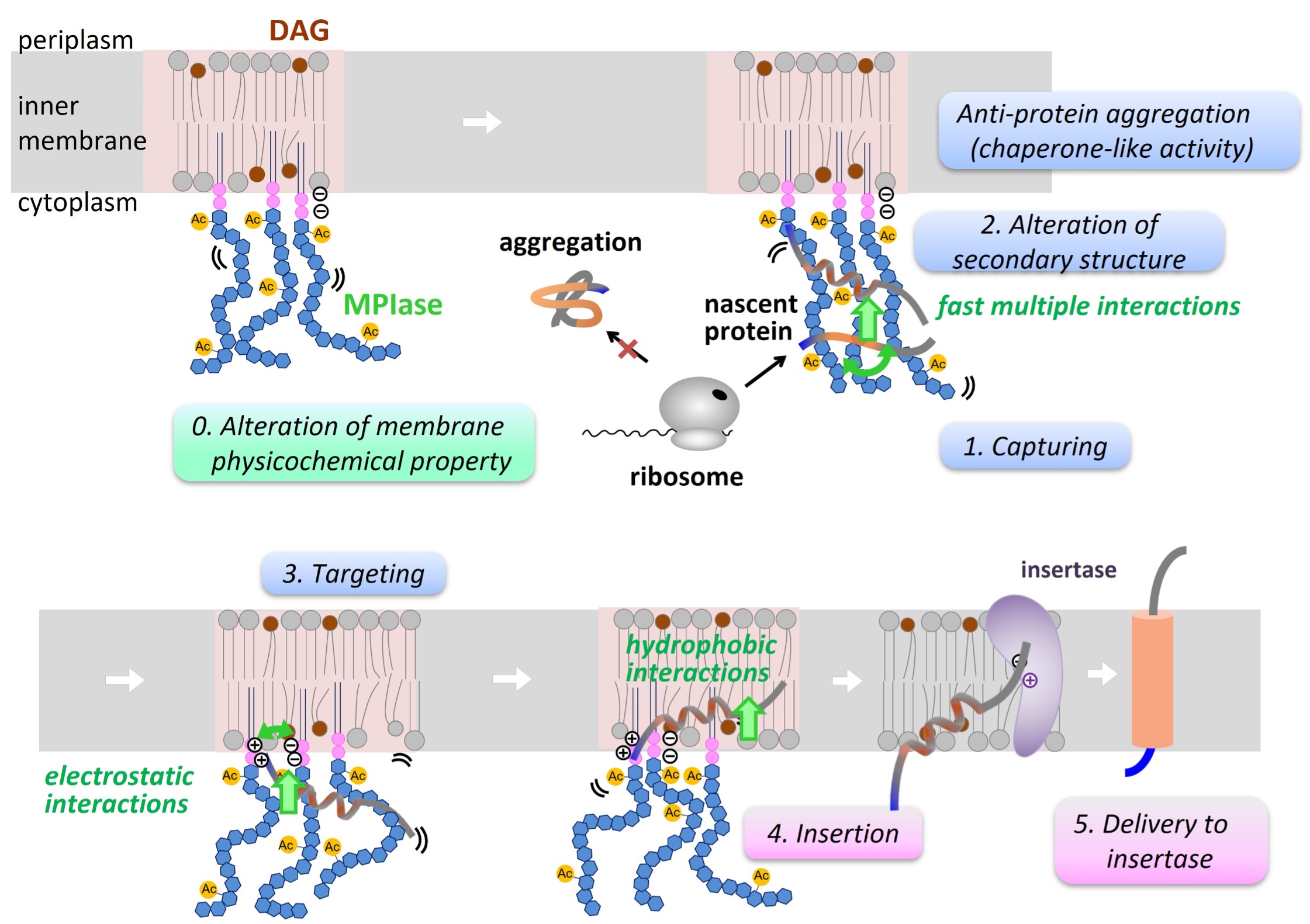
We plan to synthesize intricately structured MPIase analogs and analyze their interactions, including those with membrane integration-related proteins, to gain more insight into the membrane integration mechanism. The elucidation of the action mechanisms of MPIase will reveal the functions of minor membrane components and will contribute to the creation of new functional molecules.
3. Elucidation of the Physiological Mechanism of Plant Specialized Metabolites
Plant specialized metabolites are attracting attention as functional nutraceuticals that help improve lifestyle diseases, and their mechanisms of action at the molecular level in human body have been extensively studied in the last decades. However, little is known about their original function in plants. Therefore, we tackle the investigation of the physiological mechanisms of plant specialized metabolites by taking advantage of molecular biological or plant genetic methodologies in addition to a chemical strategy.
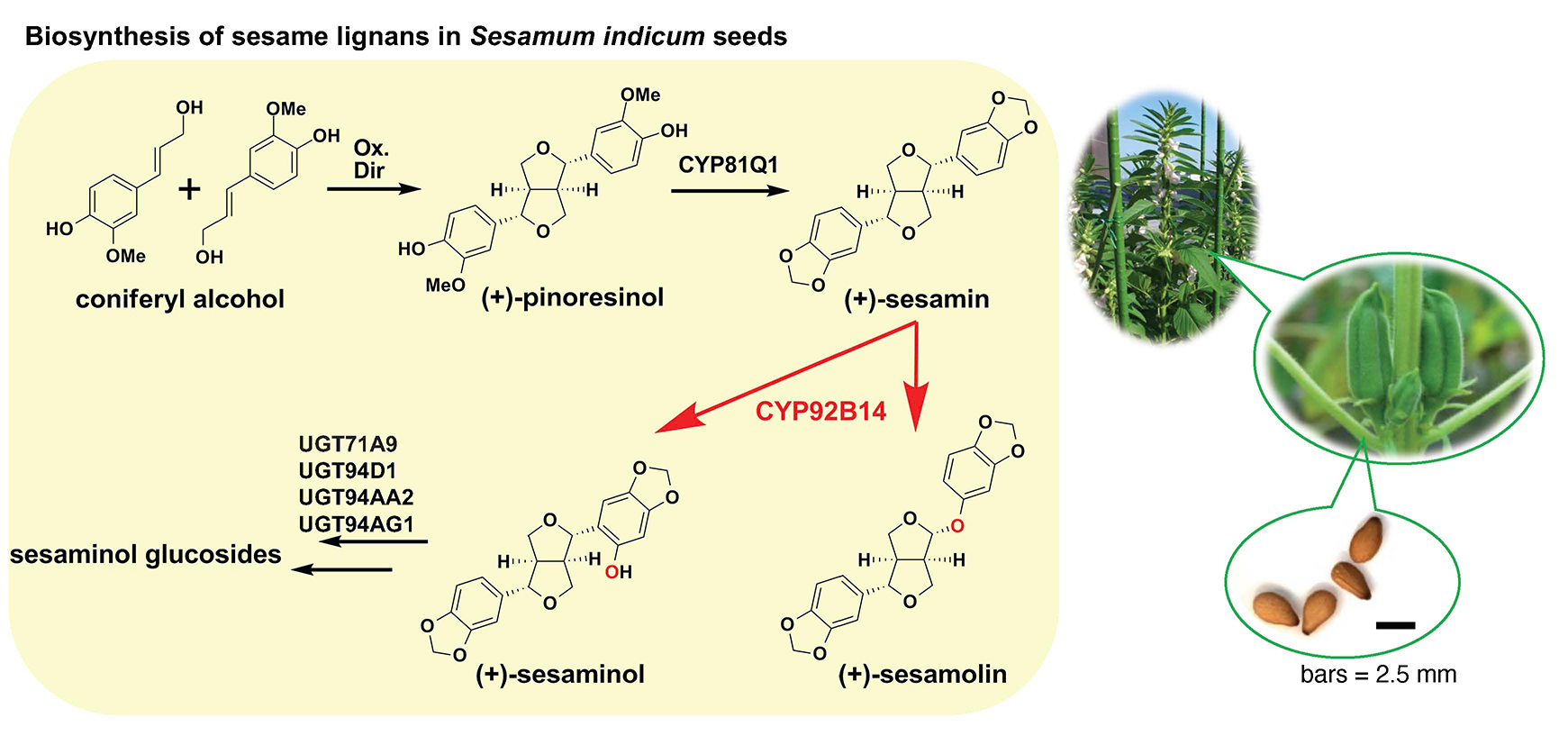
Elucidation of the Physiological Role of Sesame Lignans and Their Biosynthetic Pathway
We are currently focusing on sesamin and sesamolin, the two major lignans, a group of specialized metabolites that are found in seeds of sesame (Sesamum indicum) and exhibit anti-oxidative and health-promoting activities. In order to uncover the biological relevance of sesamin to sesame plants themselves, we performed an affinity screening using sesamin probes to reveal a candidate factor in sesame seeds, finding that sesamin specifically binds to a sterol-linked dehydrogenase. Further analyses with transgenic Aarbidopsis over-expressing the dehydrogenase gene as well as spectroscopic techniques support the view that sesamin directly binds to the binding protein.
Additionally, we are elucidating the biosynthetic pathway for sesame lignans. Although the biosynthesis of sesamin has been reported, the biosynthetic pathway for sesamolin and sesaminol glycosides, which are the major sesame lignans accumulated in sesame seeds in addition to sesamin, has not been clarified yet. We analyzed the enzymatic reaction using yeast harboring the sesamolin synthesis candidate gene (CYP92B14) in S. indicum obtained through a genetic analysis. Our results indicate that CYP92B14 protein (SiCYP92B14) produces sesamolin and sesaminol via multiple reaction schemes using sesamin as a substrate. Additionally, we recently functionally characterized SrCYP92B14 as a SiCYP92B14 orthologue from Sesamum radiatum, a wild sesame species that accumulates a sesaminol derivative sesangolin. Functional analyses suggested that the bimodal oxidation mechanism of sesamin might be widespread across Sesamum spp. and that the difference in the catalytic properties of CYP92B14 is likely to key to define the molar ratio of sesaminol- and sesamolin-derived lignans. Comparative genomics approach using various wild sesame species and cultivated sesame that exhibit distinct profiles in lignan accumulation should shed light on physiological functions of sesame lignans.
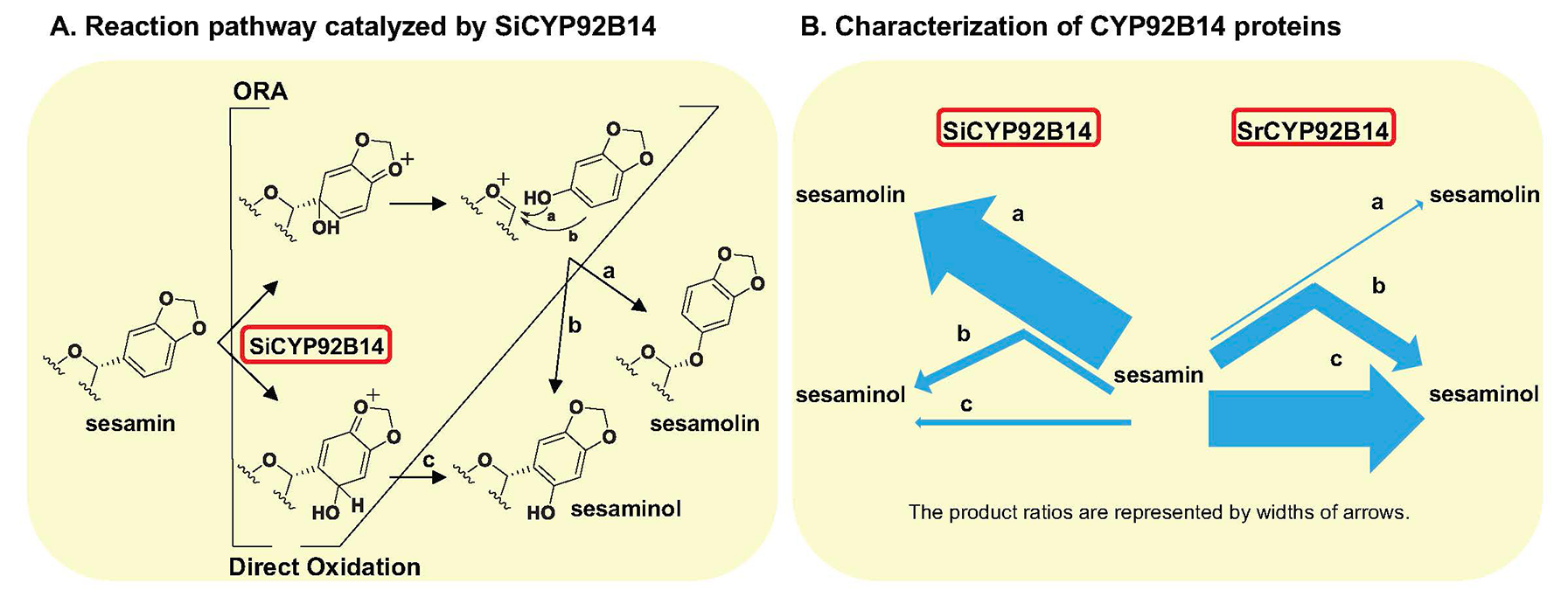
B. The reaction characteristics of SrCYP92B14, a SiCYP92B14 orthologue from the wild sesame species Sesamum radiatum, differ from those of SiCYP92B14 because of differences in amino acid residues in the substrate binding site.
4. Identification of Bioactive Natural Compounds
Natural products with potent bioactivities have attracted much attention in areas such as drug discovery, food science, and cell physiology; however, most of their biosynthetic mechanisms and functions remain unclear. We believe that unveiling these functions will deepen our understanding of the relationships between ecosystems and bioactive secondary metabolites. However, even if a natural product can be isolated, its structure is complicated and the yield is low in many cases. To solve these problems, we have utilized advanced analytical instruments such as Orbitrap MS and an 800 MHz NMR spectrometer equipped with a cryogenic probe. We are currently working on improving both the performance of these instruments and the measurement method to enable the structural elucidation of compounds with very limited availability or with complicated structures.
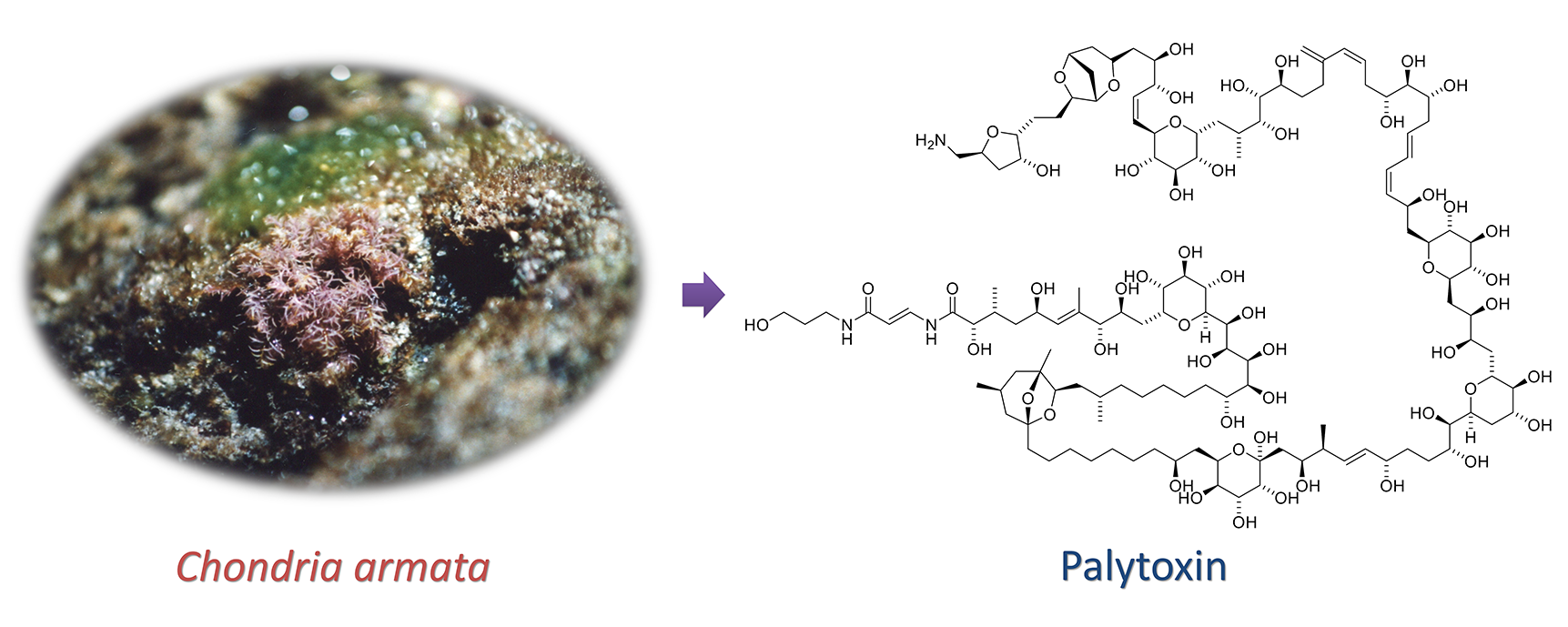
(1) Structure Determination of Palytoxins from Red Alga
Food poisoning is often caused by secondary metabolites produced by marine organisms. Consequently, the analysis of trace components in marine environments and the identification of the producers are essential not only from the viewpoint of scientific interest but also from a social perspective.
Palytoxin is one of the most toxic compounds known and exhibits an extremely large and complex structure. It was first isolated from a zoanthid, Palythoa spp., and has been found in several marine organisms such as fish and crabs. Significantly, we isolated palytoxin from marine plants. Our analyses of palytoxin and its analogs will shed some light on their biosynthesis and concentration in marine ecosystems.
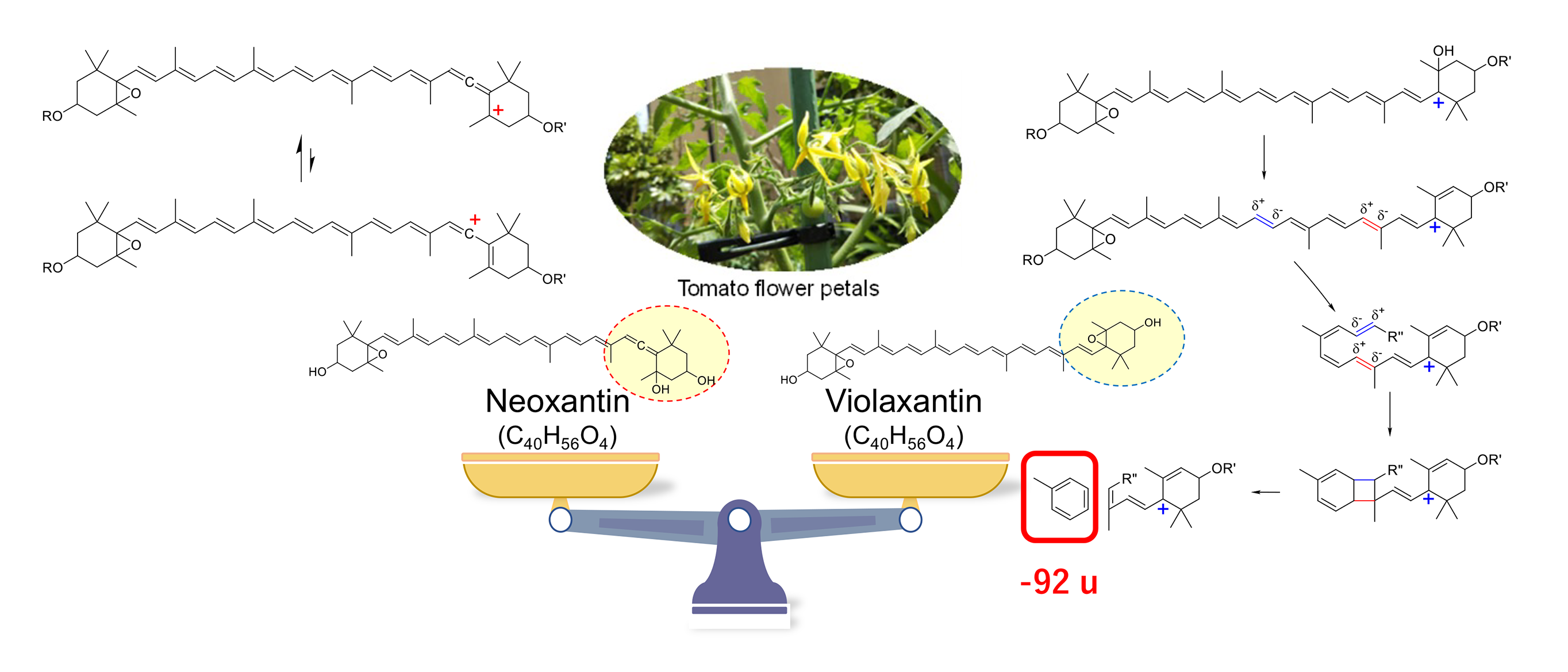
(2) Rapid Identification of Carotenoid Isomers in Yellow Petals Using Liquid Chromatography-Mass Spectrometry
Liquid chromatography-photodiode array-mass spectrometry (LC-PDA-MS) is a useful method for analyzing complex metabolites in chemotaxonomy, which involves the phylogenetic classification of plants based on the analysis of the composition of secondary metabolites and their biosynthetic pathways. However, some secondary metabolites have structural isomers that cannot be distinguished using mass numbers and absorption spectra.
In the LC-MS/MS fragmentation of carotenoids, we found differences in the intensity of 92 u loss product ions due to the stability of the carbocation in the reaction intermediate. Using this method, we rapidly discriminated between violaxanthin and neoxanthin esters, which are carotenoid pigments possessing the same molecular formula contained in yellow petals (tomato petals). We plan to develop methods for the rapid identification of metabolites on the basis of absorption spectra, mass numbers, and MS/MS fragment ion analysis using LC-PDA-MS/MS.